Isolation of DNA aptamers for herbicides under varying divalent metal ion concentrations
RESEARCH REPORT
![]()
ISSN: 2514-3247
Aptamers (2018), Vol 2, 82-87
Published online: 11 December 2018
Full Text (PDF ~388kb) (TeSelle-Supplementary Information ~264kb) | (PubMed Central Record HTML) | (PubMed) | (References)
Erienne K TeSelle and Dana A Baum*
Department of Chemistry, Saint Louis University, 3501 Laclede Avenue, St Louis, Missouri, 63013, USA
*Correspondence to: Dana Baum, Email: dana.baum@slu.edu, Tel: +1 314 977 2842, Fax: +1 314 977 2521
Received: 09 October 2018 | Revised: 10 December 2018 | Accepted: 11 December 2018
© Copyright The Author(s). This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0). This license permits non-commercial use, distribution and reproduction of this article, provided the original work is appropriately acknowledged, with correct citation details.
ABSTRACT
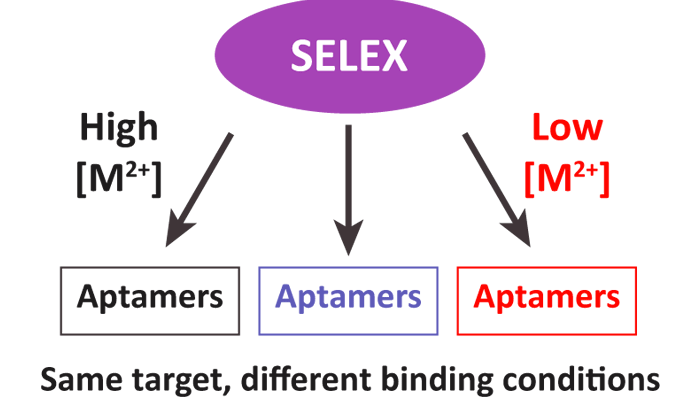
Functional nucleic acids, including aptamers and deoxyribozymes, have become important in a variety of applications, particularly sensors. Aptamers are useful for recognition because of their ability to bind to targets with high selectivity and affinity. They can also be paired with deoxyribozymes to form signaling aptazymes. These aptamers and aptazymes have the potential to significantly improve the detection of small molecule pollutants, such as herbicides, in the environment. One challenge when developing aptazymes is that aptamer selection conditions can vary greatly from optimal deoxyribozyme reaction conditions. Aptamer selections commonly mimic physiological conditions, while deoxyribozyme selections are conducted under a wider range of divalent metal ion conditions. Isolating aptamers under conditions that match deoxyribozyme reaction conditions should ease the development of aptazymes and facilitate the activities of both the binding and catalytic components. Therefore, we conducted in vitro selections under different divalent metal ion conditions to identify DNA aptamers for the herbicides atrazine and alachlor. Conditions were chosen based on optimal reaction conditions for commonly-used deoxyribozymes. Each set of conditions yielded aptamers that were unrelated to aptamers identified under other selection conditions. No particular set of conditions stood out as being optimal from initial binding analysis. The best aptamers bound their target with high-micromolar to low-millimolar affinity, similar to the concentrations used during the selection proceduresregulatory guidelines. Our results demonstrate that different metal ion concentrations can achieve the common goal of binding to a particular target, while providing aptamers that function under alternate conditions.
KEYWORDS: aptamer, SELEX, in vitro selection, herbicides, atrazine, alachlor