Chemically “barbed” aptamers selected from a base-modified RNA library
RESEARCH ARTICLE
![]()
ISSN: 2514-3247
Aptamers (2018), Vol 2, 74-81
Published online: 07 December 2018
Full Text (PDF ~2421kb) (MacPherson-Supplementary Information ~1704kb) | (PubMed Central Record HTML) | (PubMed) | (References)
Iain S MacPherson*1,2 and Tim Liedl1
1Department of Physics, Ludwig Maximilian University, Munich, Bavaria, 80539, Germany
2Department of Tropical Medicine, Medical Microbiology and Pharmacology, University of Hawaii, Honolulu, Hawaii, 96813, USA
*Correspondence to: Ian MacPherson, Email: iain.macpherson@hawaii.edu, Tel: 808 377 0878, Fax: 808 692 1677
Received: 28 August 2018 | Revised: 19 November 2018 | Accepted: 29 November 2018
© Copyright The Author(s). This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0). This license permits non-commercial use, distribution and reproduction of this article, provided the original work is appropriately acknowledged, with correct citation details.
ABSTRACT
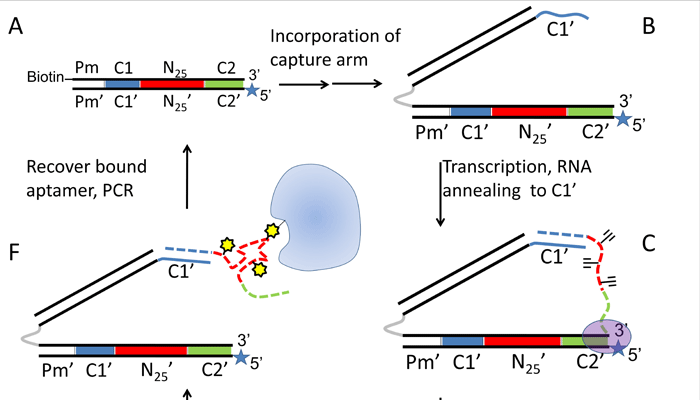
Aptamers are capable of binding to target proteins with high affinity and specificity. Recent technologies have focused on increasing the chemical diversity of libraries for the isolation of aptamers with improved binding. For this, DNA has been the nucleic acid of choice, in part due to the ability of certain DNA polymerases to accommodate modified DNA bases. Success with base-modified RNA libraries has been rare to date, despite potential advantages of RNA aptamers over DNA aptamers. Here we further expand the range of base modification in RNA aptamers by a process called RNA-SELMA (SELection with Modified Aptamers). In this process, RNA is displayed by the DNA that encodes it, enabling the selection of post-transcriptionally modified RNA libraries and amplification of their encoding DNA, thus circumventing the need for reverse transcription. In this report, we show that chemical attachment of N-hydroxysuccinimidyl esters to DNA-displayed RNA libraries enables the selection of aptamers capable of bioconjugation with a target protein, streptavidin, at low nanomolar concentration with high specificity.
KEYWORDS: SELEX, mod-SELEX, modified aptamer, RNA-SELMA, covalent aptamer, ethynyluridine