Riboswitches and synthetic aptamers: a head-to-head comparison
REVIEW ARTICLE
![]()
ISSN: 2514-3247
Aptamers (2018), Vol 2, 1-10
Published online: 04 Feb 2018
Full Text (PDF~906kb) (Supplementary Table S1 ~29kb) | (PubMed Central Record HTML) | (PubMed) | (References)
Amal Moh Awwad1 and Maureen McKeague2,*
1Institute of Biochemistry, Carleton University, 1125 Colonel By Drive, Ottawa, ON, Canada
2Department of Health Sciences and Technology, ETH Zürich, Schmelzbergstrasse 9, 8092 Zürich, Switzerland
*Correspondence to: Maureen McKeague, E-mail: maureen.mckeague@hest.ethz.ch
Received: 14 November 2017 | Revised: 25 January 2018 | Accepted: 04 February 2018
© Copyright The Author(s). This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0). This license permits non-commercial use, distribution and reproduction of this article, provided the original work is appropriately acknowledged, with correct citation details.
ABSTRACT
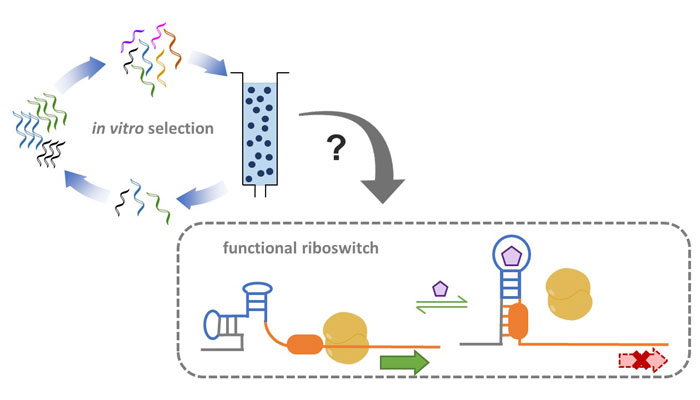
Nucleic acid aptamers are unique molecular structures used for binding to diverse targets. There is a major challenge in adapting in vitro-selected RNA aptamers for building in vivo RNA devices that control cell function. In contrast, their natural nucleic acid counterparts, riboswitches, were deliberately evolved for efficient gene regulation and cellular programming. Encoded within cells, riboswitches exploit a natural aptamer module to bind to an intracellular small molecule target enabling regulation of fundamental metabolic pathways. Here, we review several key features of natural riboswitches that may account for their function in the cellular environment. We compared these features to those of in vitro selected RNA aptamers that bind to small molecule targets. Our analysis revealed that the aptamer structure and magnesium-dependence might be the largest contributors to failed synthetic RNA devices. Thus, we make several suggestions for forthcoming aptamer selections, which may improve the success of synthetic RNA design, implementation into cells, and ultimately expand their applications.
KEYWORDS: Aptamer, riboswitch, secondary structure, binding affinity, small molecule, metabolite