MINI-REVIEW
![]()
ISSN: 2514-3247
Aptamers (2018), Vol 2, 55-63
Published online: 24 August 2018
Full Text (PDF ~1745kb) | (PubMed Central Record HTML) | (PubMed) | (References)
Anastasia A Novoseltseva1,2,*, Elena G Zavyalova1,2, Andrey V Golovin2,3, Alexey M Kopylov1,2
1Department of Chemistry, Lomonosov Moscow State University, Moscow, Russia
2Apto-Pharm Ltd, Moscow, Russia
3Department of Bioengineering and Bioinformatics, Lomonosov Moscow State University, Moscow, Russia
*Correspondence to: Anastasia Novoseltseva, Email: novoseltsevaaa@gmail.com, Tel: +749 59393149, Fax: +749 59393181
Received: 04 May 2018 | Revised: 12 July 2018 | Accepted: 23 August 2018
© Copyright The Author(s). This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0). This license permits non-commercial use, distribution and reproduction of this article, provided the original work is appropriately acknowledged, with correct citation details.
ABSTRACT
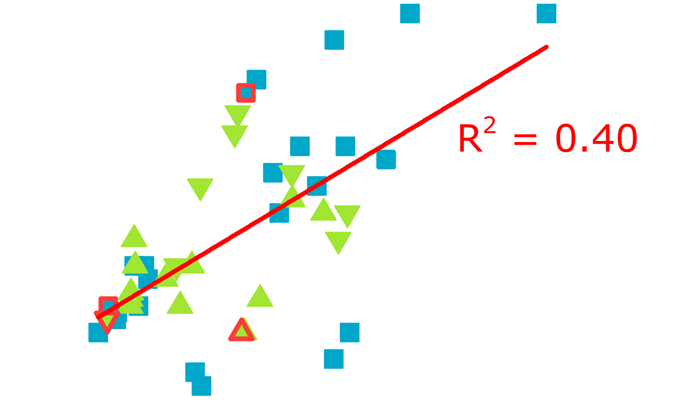
A total of forty-five X-ray structures of aptamer–protein complexes have been resolved so far. We analysed a large dataset using common aptamer parameters such as the type of nucleic acid, aptamer length and presence of chemical modifications, and the various parameters of complexes such as interface area, number of polar contacts and Gibbs free energy change. We did not find correlation between Gibbs free energy change, interface area, or number of polar contacts. The elements of the dataset with heterogeneous parameters were clustered, providing a possibility to reveal structure–affinity relationship, SAR. Complexes with DNA aptamers and RNA aptamers had the same characteristics. Presence of aptamer modifications within the interface decreased Gibbs free energy change. Furthermore, a correlation between Gibbs free energy change and the interface area of complexes with modified aptamers was found. We also attempted to compare SAR for aptamer–protein complexes with antibody–protein SARs.
KEYWORDS: X-ray three-dimensional structure, aptamer, aptamer–protein complex, polar contact, interface area, structure–affinity relationship, SAR