MEETING REVIEW
![]()
ISSN: 2514-3247
Aptamers (2018), Vol 2, 11-14
Published online: 12 Feb 2018
Full Text (PDF ~214kb) | (PubMed Central Record HTML) | (PubMed) | (References)
Justin Henri1, Maureen McKeague2, Philip E Johnson3, Beatrix Suess4, Yoshikazu Nakamura5,6, Marit Nilsen-Hamilton7,8, Fernando Pastor9, Ulrich Hahn10, David Bunka11, Sarah Shigdar1,12
1School of Medicine Deakin University, Geelong, Victoria 3128, Australia
2Department of Health Sciences
and Technology, ETH Zürich, Schmelzbergstrasse 9, 8092 Zürich, Switzerland
3Department of Chemistry and Centre for Research on Biomolecular Interactions, York University, Toronto, Ontario, Canada, M2R 1A1
4Department of Biology, Technical University Darmstadt, Schnittspahnstr 10, 64287 Darmstadt, Germany
5RIBOMIC Inc, Tokyo 108-0071, Japan
6The Institute of Medical Science, University of Tokyo, Tokyo 108-8639
7Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA 50011, USA
8Aptalogic Inc., Ames, IA, United States
9Molecular Therapy Program, Center for Applied Medical Research (CIMA), Foundation for Applied Medical Research (FIMA), Pamplona, Spain
10Institute for Biochemistry and Molecular Biology, Chemistry Department, University of Hamburg, Martin-Luther- King-Platz 6, D-20146 Hamburg, Germany
11Aptamer Group Ltd, Bio Centre, Innovation Way, Heslington, York, UK
12Centre for Molecular and Medical Research, Deakin University, Geelong, Victoria 3128, Australia
*Correspondence to: Sarah Shigdar, Email: sarah.shigdar@deakin.edu.au
Received: 31 January 2018 | Revised: 12 February 2018 | Accepted: 12 February 2018
© Copyright The Author(s). This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0). This license permits non-commercial use, distribution and reproduction of this article, provided the original work is appropriately acknowledged, with correct citation details.
The 4th annual symposium of the International Society on Aptamers, Aptamers 2017, was held in Oxford on the 11th and 12th April and was well attended, with presenters from Europe (Spain, Germany, Austria, and UK), North and South America, Asia, and Australasia presenting on a diverse range of topics, from enhancing SELEX, to diagnostic applications such as lateral flow devices or medical imaging, to therapeutic applications such as drug delivery. The conference was split into six sections in total, covering chemical modifications, disease, analysis/diagnostics, tools/selection/design, riboswitches, and computation with over 50 oral and poster presentations. The conference started with a welcome from both the Symposium Chair, Professor Dr Ulrich Hahn (University of Hamburg, Germany) and the President of the International Society on Aptamers, Dr Sarah Shigdar (Deakin University, Australia).
CHEMICAL MODIFICATIONS OF APTAMERS
Aptamers are developing for a number of applications and they have demonstrated high potential for therapeutic applications. However, some aptamers are prone to rapid nuclease degradation. Therefore, in order to develop aptamers for clinical applications, some modification of the nucleotides are required (Shigdar et al, 2013). Professor Philip Johnson (York University, Canada) chaired this first session where we heard from a number of leading experts in the field. Professor Jesper Wengel (University of Southern Denmark, Denmark), the co-inventor of locked nucleic acids, started off the first session by providing an update on the use of Phusion® High Fidelity DNA polymerase and engineered polymerases as a way forward for SELEX using LNAs (Lou et al, 2017b), definitely something that has potential for better bioavailability of aptamers in therapeutic applications. Professor Wengel also discussed the use of LNAs and unlocked nucleic acids as antisense or RNAi drug candidates before presenting results on how to build artificial protein mimics through the combination of oligonucleotides and short peptide sequences (Lou et al, 2017a). Dr Xianbin Yang (AM Biotechnologies, USA) from AM Biotechnologies, then presented information on X-aptamers which are developed using a microbead based single-cycle discovery process and does not rely on PCR amplification, allowing a variety of chemical modifications to be incorporated (Lokesh et al, 2017). Dr Dan Schneider (SomaLogic, USA) from SomaLogic was the third presenter in this session and he talked about enhancing chemical diversity with multiple pyrimidine modifications. These modifications can create novel intramolecular motifs leading to improved aptamer affinities (Gawande et al, 2017).
DISEASE
One of the areas in which aptamers are showing a lot of potential is the area of theranostics. Aptamers bind to their target through complex interactions involving hydrogen bonding, van der Waals forces, hydrophobic, and electrostatic interactions. In this manner, aptamers can act as agonists or antagonists, or through receptor mediated endocytosis, the aptamers can be used for targeted drug delivery (Macdonald et al, 2018). Additionally, aptamers can demonstrate higher sensitivity in diagnostic assays, especially in the case of complex diseases. The second session, chaired by Professor Dr Beatrix Suess (Technical University Darmstadt, Germany), discussed various different ways aptamers are being used to effectively target and treat, or more sensitively diagnose disease. Dr Greg Penner (NeoNeuro SAS, Paris, France) started the session with a discussion of aptamer theranostics for Alzheimer’s disease. Dr Penner’s team preselected the randomised library by injecting the library into the tail vein of mice and then collecting only the aptamers that has passed into the brain. They then performed aptamer selection on human blood from Alzheimer’s patients to pick out targets that could be used as a diagnostic panel. Their panel was effective in stratifying patients into early, middle and late stage Alzheimer’s disease and they are now investigating this Aptamarker AD fingerprint for earlier diagnosis of disease (Lecocq et al, 2018). Next was Professor Dr Ulrich Hahn (University of Hamburg, Germany) and our Symposium Chair, who presented data on selectin and integrin aptamers as a means of preventing metastasis. Both of these aptamers were shown to have an anti-adhesive effect on cancer cells though truncation of the integrin binding aptamer had a detrimental effect on the function of the aptamer (Berg et al, 2016). Professor Yoshikazu Nakamura (Ribomic Inc, Japan) presented on a fibroblast growth factor 2 (FGF-2) binding aptamer that can restore bone growth in Achondroplasia (ACH) and is now gearing up for clinical trials. This aptamer has restored body weight and length in ACH transgenic mice and has an excellent profile in rat and monkey plasma. They are now looking at it in age-related macular degeneration (Jin et al, 2016). Professor Fernando Pastor (Foundation for Applied Medical Research, Spain) presented data on generating a co-stimulatory agonistic aptamer. This aptamer, which bound to both CD28 and multidrug resistant protein 1 (MRP-1), inhibited growth of tumours in mice (Soldevilla et al, 2016). Last in this session was Dr Sarah Shigdar (Deakin University, Australia) who presented data on a bifunctional aptamer capable of both crossing the blood brain barrier from systemic circulation and specifically targeting cancer cells in the brain. This aptamer has been tested in vitro and in vivo and shows promise for future targeted drug delivery (Macdonald et al, 2017).
ANALYSIS/DIAGNOSTICS
As in the previous session, where there was a discussion relating to sensitive diagnosis of Alzheimer’s disease by means of a panel of aptamers, this session reviewed some of the developing aspects of nanotechnology and point-of-care testing. Chaired by Professor Yoshikazu Nakamura, the session started with some exciting work presented by Professor Anthony Cass (Imperial College London, UK). Professor Cass discussed his diagnostics tool, dual recognition element lateral flow assay (DREFLA), to differentiate specific influenza strains from other similar strains (Le et al, 2017). Next Dr Julian Tanner (University of Hong Kong, PR China) presented data on using aptamers for malaria diagnostics, and the APTEC test for Plasmodium falciparum. He has also developed ‘origami boxes’ which ‘opens’ when the target is present, allowing for better point of care testing (Dirkzwager et al, 2015; Godonoga et al, 2016). Last in this session was Professor Victoria Calzada (Universidad de la República, Montevideo-Uruguay) who has linked technetium-99m and gallium-67 to an aptamer, sgc-8, through the chelators HYNIC and DOTA, and which showed good biodistribution and pharmacokinetics (Calzada et al, 2017).
TOOLS/SELECTION/DESIGN (PART I)
As technology is advancing rapidly, so too is the ability to develop aptamers more rapidly, or for advanced applications. As there are a number of exciting developments in this area, this topic was split across two sessions. The first part of this session, and the last for day 1 was chaired by Professor Marit Nilsen-Hamilton (Iowa State University, USA). Dr David Bunka (Aptamer Group Ltd, UK) started the session with a discussion on ‘Aptabind’ and their industrial application of protein purification. Their patented technology can be ‘tuned’ to the process to ensure that the process is not only efficient, but is also capable of purifying intact functional proteins. Next up was Dr Duncan Borthwick (Dynamic Biosensors GmbH, Germany) who has developed an electroswitchable biosurface to measure accurate binding kinetics of aptamers. Following this, Dr Sean Dembowski (University of Minnesota–Twin Cities, USA) discussed capillary electrophoresis as a means of enhancing the SELEX process for membrane proteins (Dembowski and Bowser 2017). Last of the session on day 1 was Mr John Goertz (University of Maryland, USA) who discussed a novel point-of-care device that uses a peroxidyme-based system allowing for assay automation (Goertz and White, 2017).
TOOLS/SELECTION/DESIGN (PART II)
Starting off day 2 and continuing the Tools/Selection/Design session, and chaired by Professor Fernando Pastor, was Dr Terry Steele (Nanyang Technological University, Singapore) who discussed the applications of multiple use fluorescent aptasensors, which could be used for rapid detection in real time field samples and effluents (Zhou et al, 2018). Dr Meltem Avci-Adali (University Hospital Tuebingen, Germany) discussed the use of aptamer functionalised hydrogels loaded with VEGF aptamers to capture endothelial cells to provide nutrients and oxygen delivery to cells (Guan et al, 2017). Last in this session was Professor Philip Johnson who discussed the complexities of two-site binding of the cocaine-binding aptamer, as well as the ATP-binding aptamer (Neves et al, 2017).
RIBOSWITCHES
The penultimate session focused on riboswitches and was chaired by Professor Ulrich Hahn. Riboswitches are RNA elements within an RNA transcript that modulate expression of proteins encoded by messenger RNA (Garst et al, 2011). Professor Beatrix Suess discussed how aptamers can be applied as engineered riboswitches, providing several exciting recent examples from her research group (Berens et al, 2015). For example, the Suess group has developed strategies to control exon skipping for splicing in cells, as well as tuneable regulation of cell apoptosis. Dr Florian Groher (Technical University Darmstadt, Germany) then presented his work on the de novo selection and subsequent in vivo screening of a new synthetic riboswitch responsive to the antibiotic ciprofloxacin ( Groher and Suess, 2016; Groher et al, 2018). Both presentations emphasized the challenges in developing riboswitches from in vitro selected aptamers, and highlighted the need for improved in vivo screening systems following conventional SELEX. Dr Carlos Penedo (University of St Andrews, UK) presented elegant and insightful studies using single-molecule FRET for the structural analysis of the aptamer-expression platforms in riboswitch sequences (Blouin et al, 2015; Perez-Gonzalez et al, 2016). Completing the session on riboswitches, Professor Mario Mörl (Leipzig University, Germany), discussed some of the benefits and challenges of the modular composition of a standard riboswitch (Etzel and Mörl, 2017).
COMPUTATION
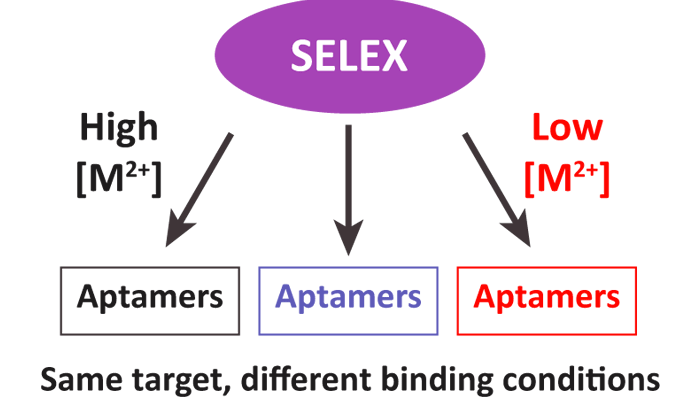
The final session, chaired by Dr David Bunka, focused on computational methods for analysing or selecting aptamers. This is a growing area of research, given the new applications available for in silico analysis of aptamers for both structure determination and selection. First, Dr Muslum Ilgu (Aptalogic Inc, USA) discussed his combined approach, using computational models and biochemical interrogation to determine aptamer 3D structure. Specifically, MC-Sym was used to predict a number of structures in silico. Next, 2-aminopurine (2-AP)-substituted aptamers were generated and analysed. With this modification, changes in fluorescence represented regions of structural flexibility and facilitated a rapid, and relatively inexpensive method for determining aptamer structure. Next, Dr Greg Penner (NeoVentures Biotechnology Inc, Canada) presented again, this time representing NeoVentures Biotechnology. At NeoVentures, they have developed an analysis of SELEX progression using next generation sequencing at each round of selection. Dr Penner spoke of SELEX as being a combination of math and chemistry. For example, as expected, as a selection experiment proceeds, the complexity of the pool decreases; however, NeoVentures tends to observe similar sequences beginning at rounds 7-10, and it is the trajectory throughout the selection experiments that is important when isolating/finding the best aptamer sequences. In the final talk of the conference, Dr Maureen McKeague (ETH Zürich, Switzerland) discussed her analysis of the past 23 years of SELEX experiments. Her analysis revealed that the best selection experiments (e.g., those with the highest affinity aptamers) were done at 37 degrees Celsius, with low magnesium concentrations, and using efficient methods for separating PCR double-stranded products (McKeague et al, 2015).
CONCLUSION
The conference concluded with Professor Dr Ulrich Hahn providing closing remarks prior to wishing everyone a safe journey home and a return for Aptamers 2018, which is the 5th Aptamers Symposium on 11th and 12th April 2018.
ACKNOWLEDGEMENTS
We are grateful to our sponsors for their generous support, including Aptamer Group, Ribomic Inc, SciEx, Somalogic, ChemGenes Corporation, NeoVentures, Library Publishing Media, and INSOAP.
REFERENCES
Berens C, Groher F and Suess B. 2015. RNA aptamers as genetic control devices: The potential of riboswitches as synthetic elements for regulating gene expression. Biotechnology Journal, 10, 246-257.
Berg K, Lange T, Mittelberger F, Schumacher U and Hahn U. 2016. Selection and Characterization of an alpha6beta4 Integrin blocking DNA Aptamer. Mol Ther Nucleic Acids, 5, e294.
Blouin S, Craggs TD, Lafontaine DA and Penedo JC 2015, ‘Functional Studies of DNA-Protein Interactions Using FRET Techniques’, in BP Leblanc & S Rodrigue (eds), DNA-Protein Interactions: Principles and Protocols, Springer New York, New York, NY, pp. 115-141.
Calzada V, Moreno M, Newton J, et al. 2017. Development of new PTK7-targeting aptamer-fluorescent and -radiolabelled probes for evaluation as molecular imaging agents: Lymphoma and melanoma in vivo proof of concept. Bioorg Med Chem, 25, 1163-1171.
Dembowski SK and Bowser MT. 2017. Microfluidic methods for aptamer selection and characterization. Analyst, 143, 21-32.
Dirkzwager RM, Kinghorn AB, Richards JS and Tanner JA. 2015. APTEC: aptamer-tethered enzyme capture as a novel rapid diagnostic test for malaria. Chem Commun (Camb), 51, 4697-4700.
Etzel M and Mörl M. 2017. Synthetic Riboswitches: From Plug and Pray toward Plug and Play. Biochemistry, 56, 1181-1198.
Garst AD, Edwards AL and Batey RT. 2011. Riboswitches: Structures and mechanisms. Cold Spring Harb Perspect Biol, 3, a003533.
Gawande BN, Rohloff JC, Carter JD, et al. 2017. Selection of DNA aptamers with two modified bases. Proc Natl Acad Sci U S A, 114, 2898-2903.
Godonoga M, Lin TY, Oshima A, et al. 2016. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci Rep, 6, 21266.
Goertz JP and White IM. 2017. Peroxidyme-Amplified Radical Chain Reaction (PARCR): Visible Detection of a Catalytic Reporter. Angew Chem Int Ed Engl, 56, 13411-13415.
Groher F, Bofill-Bosch C, Schneider C, et al. 2018. Riboswitching with ciprofloxacin—development and characterization of a novel RNA regulator. Nucleic Acids Research, gkx1319-gkx.
Groher F and Suess B. 2016. In vitro selection of antibiotic-binding aptamers. Methods, 106, 42-50.
Guan X, Avci-Adali M, Alarçin E, et al. 2017. Development of hydrogels for regenerative engineering. Biotechnology Journal, 12, 1600394.
Jin L, Nonaka Y, Miyakawa S, Fujiwara M and Nakamura Y. 2016. Dual Therapeutic Action of a Neutralizing Anti-FGF2 Aptamer in Bone Disease and Bone Cancer Pain. Mol Ther, 24, 1974-1986.
Le TT, Chang P, Benton DJ, et al. 2017. Dual Recognition Element Lateral Flow Assay Toward Multiplex Strain Specific Influenza Virus Detection. Anal Chem, 89, 6781-6786.
Lecocq S, Spinella K, Dubois B, et al. 2018. Aptamers as biomarkers for neurological disorders. Proof of concept in transgenic mice. PLoS One, 13, e0190212.
Lokesh GL, Wang H, Lam CH, et al. 2017. X-Aptamer Selection and Validation. Methods Mol Biol, 1632, 151-174.
Lou C, Christensen NJ, Martos-Maldonado MC, et al. 2017a. Folding Topology of a Short Coiled-Coil Peptide Structure Templated by an Oligonucleotide Triplex. Chemistry, 23, 9297-305.
Lou C, Samuelsen SV, Christensen NJ, Vester B and Wengel J. 2017b. Oligonucleotides Containing Aminated 2′-Amino-LNA Nucleotides: Synthesis and Strong Binding to Complementary DNA and RNA. Bioconjug Chem, 28, 1214-1220.
Macdonald J, Henri J, Goodman L, et al. 2017. Development of a Bifunctional Aptamer Targeting the Transferrin Receptor and Epithelial Cell Adhesion Molecule (EpCAM) for the Treatment of Brain Cancer Metastases. ACS Chem Neurosci, 8, 777-784
Macdonald J, Henri J, Roy K, et al. 2018. EpCAM Immunotherapy versus Specific Targeted Delivery of Drugs. Cancers (Basel), 10, 19
McKeague M, McConnell EM, Cruz-Toledo J, et al. 2015. Analysis of In Vitro Aptamer Selection Parameters. Journal of Molecular Evolution, 81, 150-161.
Neves MAD, Slavkovic S, Churcher ZR and Johnson PE. 2017. Salt-mediated two-site ligand binding by the cocaine-binding aptamer. Nucleic Acids Research, 45, 1041-1048.
Perez-Gonzalez C, Lafontaine DA and Penedo JC. 2016. Fluorescence-Based Strategies to Investigate the Structure and Dynamics of Aptamer-Ligand Complexes. Frontiers in Chemistry, 4, 33.
Shigdar S, Macdonald J, O’Connor M, et al. 2013. Aptamers as theranostic agents: modifications, serum stability and functionalisation. Sensors (Basel), 13, 13624-13637.
Soldevilla MM, Villanueva H, Casares N, et al. 2016. MRP1-CD28 bi-specific oligonucleotide aptamers: target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget, 7, 23182-23196.
Zhou Y, Wu Y, Pokholenko O, et al. 2018. Aptamer adaptive binding assessed by stilbene photoisomerization towards regenerating aptasensors. Sensors and Actuators B: Chemical, 257, 245-255.